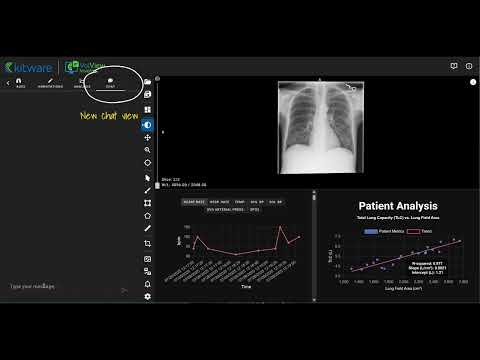
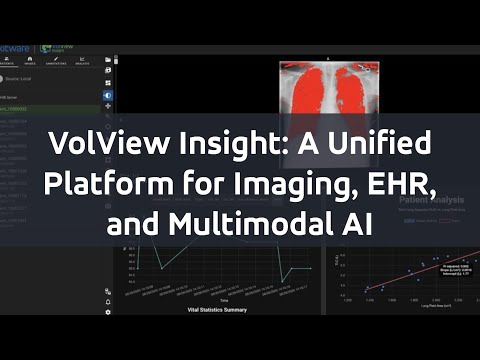
VolView Insight is an open-source platform that unifies medical imaging and clinical data into one extensible interface. It provides researchers with a realistic testbed for developing, testing, and extending multimodal models in settings that mirror clinical reality.
- 🔬 Unified access to imaging data (DICOM/DICOMWeb) and clinical records (FHIR/EHR).
- 🧠 Extensible backend for AI/ML pipelines, including deep learning inference.
- ⚡ Web-based UI built on VolView.
- 🔌 Pluggable integrations — Orthanc, HAPI on FHIR, and MedGemma are examples. Users can bring their own DICOM/FHIR servers or multimodal pipelines.
- Docker
20.10.0or later - Docker Compose v2.0 or later (included with Docker Desktop)
Note on Docker Compose command: This project uses
docker-compose(with hyphen) in all scripts. Docker Compose V2 is available in two forms:
- Standalone binary:
docker-compose(with hyphen) - common in Linux/server environments- Docker plugin:
docker compose(no hyphen) - bundled with Docker DesktopBoth refer to the same modern Docker Compose V2. Our scripts use the standalone form for maximum compatibility across different installation types. If you have Docker Desktop, both commands will work interchangeably.
The easiest way to get started is using our Docker orchestration setup:
git clone https://github.com/KitwareMedical/volview-insight.git
cd volview-insight
git submodule update --init# Setup data volumes (creates both volumes/ and volumes-db/ structures)
./scripts/setup-volumes.sh
# Create environment file from template
cp env.example .envImportant for Chat Mode (MedGemma):
To use the chat mode (conversational AI) in VolView Insight, which relies on the MedGemma model:
- Create a Hugging Face account
- Generate a Hugging Face access token (read-only permission is sufficient)
- Log in to your Hugging Face account and accept the MedGemma license terms (you must do this while logged in, or your requests will be rejected)
- Request and receive permission to use MedGemma if prompted
You must complete all these steps on the Hugging Face website before proceeding here.
# Add your Hugging Face token needed for gated public model like medgemma:
HF_TOKEN=hf_your_token_hereVolume Structure Overview: VolView Insight uses two separate volume directories:
volumes/- Source data (raw files you want to import)volumes-db/- Docker persistent volumes (database storage managed by Docker)
Directory Organization:
volumes/ # SOURCE DATA (your raw files)
├── orthanc-data/ # Raw DICOM files to be imported
│ ├── patient_[ID]/ # Patient directories by Patient ID
│ │ ├── study1.dcm # DICOM files for this patient
│ │ └── study2.dcm
│ └── dicom_metadata.json # Import metadata (optional)
└── hapi-fhir-data/ # Raw FHIR resources to be imported
├── patient_*.json # Individual FHIR patient resources
├── patients.json # Combined patient list (optional)
└── [resource_type]/ # Additional FHIR resources by type
├── condition/
├── observation/
└── ...
volumes-db/ # DOCKER VOLUMES (persistent databases)
├── orthanc-data/ # Orthanc database (auto-managed)
├── hapi-fhir-data/ # HAPI FHIR database (auto-managed)
└── model-cache/ # AI model cache (persistent)
├── segmentLungsModel-v1.0.ckpt
└── medgemma/ # Downloaded on first use
For new users cloning from GitHub:
-
Prepare your source data in the
volumes/directory structure:# Place raw DICOM files in volumes/orthanc-data/ # Place raw FHIR resources in volumes/hapi-fhir-data/
-
Start the services (creates empty databases in
volumes-db/):./scripts/start.sh
-
Import your data from source to databases:
# Auto-import from volumes/ to running servers ./scripts/auto-import-data.sh # Or import individually: python3 ./scripts/import-dicom-to-orthanc.py python3 ./scripts/import-fhir-to-hapi.py
Note on dicom_metadata.json: If you copy example data from another machine, the
dicom_metadata.jsonfile may contain absolute paths that won't work on your system. Don't worry - the import script will automatically scan thevolumes/orthanc-data/directory for all.dcmfiles if the metadata file is missing or has invalid paths.
Alternative: Direct API uploads You can also upload data directly through REST APIs:
- DICOM: Use Orthanc's REST API at http://localhost:8042
- FHIR: Use HAPI FHIR's REST API at http://localhost:3000/hapi-fhir-jpaserver/
Data Requirements:
- DICOM files: Must contain Patient ID in DICOM headers
- FHIR data: Patient resources with matching Patient IDs
- Patient matching: Both datasets should use the same Patient ID system for proper correlation
Important: The application only displays data where Patient IDs match between FHIR and DICOM/Orthanc. If a patient exists in FHIR but has no corresponding DICOM studies with the same Patient ID, or vice versa, that data will not be available in the application interface.
Volume Persistence:
- ✅
volumes-db/data persists across container restarts - ✅ AI model cache (
volumes-db/model-cache/) persists downloads ⚠️ volumes/is your source data (not modified by the application)- 🗑️ Use
docker-compose down -vto clear all databases
🔄 Data Persistence After Initial Setup: Once you've completed the initial setup and data import, future container restarts are simple and require NO re-importing:
# After initial setup, future restarts are this simple:
./scripts/stop.sh # Stop all services
./scripts/start.sh # Start all services
# Your data will be immediately available:
# ✅ All DICOM studies (preserved in Orthanc database)
# ✅ All FHIR resources (preserved in HAPI FHIR database)
# ✅ All AI models (preserved in model cache)
# ✅ No re-import needed!Important: The first time you set up the system, you need to import data once. After that, all data persists automatically across container restarts, system reboots, and Docker updates.
# Start all services (smart build detection)
./scripts/start.sh
# First run: automatically builds images
# Subsequent runs: uses existing images (fast)
# Background mode (recommended for development)
./scripts/start.sh -d # or --detach
# Then view logs: docker-compose logs -f
# Force rebuild (after dependency changes)
./scripts/start.sh --build
# Combined flags
./scripts/start.sh -d --build # background + rebuild
# Stop all services
./scripts/stop.shSmart Build Detection:
- First run: Automatically detects missing images and builds them
- Subsequent runs: Skips build for faster startup (~5 seconds vs ~2 minutes)
- Rebuild when needed: Use
--buildflag after:- Changing
package.json,pyproject.toml, or Dockerfile - Adding new dependencies
- Updating base images
- Changing
🔄 Service Restarts:
# When you make code changes, restart specific services:
docker-compose restart backend volview-insight-web
# Or restart all services (preserves data):
./scripts/stop.sh && ./scripts/start.shAccess Points:
- Frontend: http://localhost:8080
- Backend API: http://localhost:4014
- Orthanc DICOM: http://localhost:8042
- HAPI FHIR: http://localhost:3000/hapi-fhir-jpaserver/
- Orthanc CORS Proxy: http://localhost:5173
Features:
- ✅ Hot reloading with Vite dev server
- ✅ Live code updates with volume mounts
- ✅ All services orchestrated automatically
- ✅ CORS proxy for DICOM Web API access
- ✅ Environment variables pre-configured
- ✅ Persistent database volumes in
volumes-db/
Once all services are running:
- Open the application: Navigate to http://localhost:8080
- Connect to FHIR: Click "Connect" in the FHIR Patients section
- Select a patient: Choose from the loaded patient list
- View imaging studies: Patient images will appear in the DICOM section
- Explore AI features: Use the analysis modules with your medical data
Expected behavior:
- ✅ FHIR patients load with proper names
- ✅ DICOM studies match FHIR patients by Patient ID
- ✅ Patient names display correctly even if DICOM files are anonymized
- ✅ Medical images load and display in the 3D viewer
"No matching imaging studies found"
- Ensure Patient IDs exactly match between FHIR and DICOM data (case-sensitive)
- Check that both FHIR patients and DICOM studies were imported successfully
- Verify the Patient ID field exists in DICOM headers and FHIR resources
- Run
./scripts/auto-import-data.shto verify import completed successfully - Verify services are running:
docker-compose ps - Check if data exists in source directories:
ls volumes/orthanc-data/andls volumes/hapi-fhir-data/
FHIR connection errors
- Ensure HAPI FHIR server is accessible at http://localhost:3000/hapi-fhir-jpaserver/
- Check that FHIR data was imported correctly
- Restart services:
docker-compose restart
Patient names appear blank
- This can happen with anonymized DICOM datasets
- The application automatically falls back to FHIR patient names when available
- Ensure FHIR data includes patient names in the correct format
CORS errors accessing DICOM
- Development mode uses CORS proxy at http://localhost:5173
- Ensure Orthanc proxy service is running
- Check browser developer console for detailed errors
Docker volume management
- Data persists in
volumes-db/across container restarts - To clear all databases:
docker-compose down -v - To backup data: Copy
volumes-db/directory - To restore data: Replace
volumes-db/directory before starting services - AI models download once to
volumes-db/model-cache/and persist
- DICOMWeb: Replace Orthanc with your own server.
- FHIR/EHR: Replace HAPI with your own server.
- Pipelines: Extend the Python backend with your custom multimodal methods.
We welcome contributions!
- Add new pipelines or models
- Add integrations with other EHR/DICOM backends
- Report bugs and propose features
This software is provided solely for research and educational purposes. It is a proof-of-concept research platform and is not intended for clinical use.
- This software has not been reviewed or approved by the U.S. Food and Drug Administration (FDA) or any other regulatory authority.
- It must not be used for diagnosis, treatment, or any clinical decision-making.
- No warranties or guarantees of performance, safety, or fitness for medical purposes are provided.
By using this software, you acknowledge that it is for non-clinical, investigational research only.
Apache 2.0 © Kitware