diff --git a/README.md b/README.md
index 42c130c15..9cc0d73b5 100644
--- a/README.md
+++ b/README.md
@@ -125,6 +125,7 @@ Below is an overview of all content linked inside of this document repository. [
* Phase 6: Iterating on Your Closed Loop
* So you think you’re looping? Now keep up to date!
* Testing during the day
+ * Usability Considerations
* Advanced features
* Configuring Automatic Sensitivity Mode
- Resources
diff --git a/docs/Resources/wifi.md b/docs/Resources/wifi.md
new file mode 100644
index 000000000..ce4feae94
--- /dev/null
+++ b/docs/Resources/wifi.md
@@ -0,0 +1,24 @@
+###Wifi Troubleshooting
+
+#My wifi connection keeps dropping and/or I keep getting kicked out of ssh
+There is a script that you can add to your root cron that will test your connection and reset it if it is down. Here is an example that runs every two minuntes (odd minutes). You could also do it every 5 minutes or less. Note, this does not have to be for an Edison, you can set this up for a Pi, etc as well.
+
+```
+cd ~/src
+git clone https://github.com/TC2013/edison_wifi
+cd edison_wifi
+chmod 0755 /home/edison/src/edison_wifi/wifi.sh
+```
+Next, add the script to your root cron. Note this is a different cron that what your loops runs on, so when you open it don't expect to see your loop and other items you've added.
+ * Log in as root ```su root```
+ * Edit your root cron ```crontab -e```
+ * Add the following line ```1-59/2 * * * * /home/edison/src/edison_wifi/wifi.sh google.com 2>&1 | logger -t wifi-reset```
+
+#I forget to switch back to home wifi and it runs up my data plan
+You can add a line to your cron that will check to see if is avaiable and automatically switch to it if you are on a different network.
+ * Log in as root ```su root```
+ * Edit your root cron ```crontab -e```
+ * Add the following line ```*/2 * * * * ( (wpa_cli status | grep > /dev/null && echo already on ) || (wpa_cli scan > /dev/null && wpa_cli scan_results | egrep > /dev/null && udo pa_cli select_network $(wpa_clilist_networks | grep jsqrd | cut -f 1) && echo switched to && sleep 15 && (for i in $(wpa_cli list_networks | grep DISABLED | cut -f 1); do wpa_cli enable_network $i > /dev/null; done) && echo and re-enabled other networks) ) 2>&1 | logger -t wifi-select```
+
+#I am having trouble consistently connecting to my wifi hotspot when I leave the house (iPhone)
+When you turn on your hotspot it will only broadcast for 90 seconds and then stop (even if it is flipped on). So, when you leave your house you need to go into the hotspot setting screen (and flip on if needed). Leave this screen open until you see your rig has connected. It make only take a few seconds or a full minute.
diff --git a/docs/docs/Images/Pull_Request.png b/docs/docs/Images/Pull_Request.png
new file mode 100644
index 000000000..0c4d9bcb4
Binary files /dev/null and b/docs/docs/Images/Pull_Request.png differ
diff --git a/docs/docs/Resources/my-first-pr.md b/docs/docs/Resources/my-first-pr.md
index 18fb17b2e..2d41e9148 100644
--- a/docs/docs/Resources/my-first-pr.md
+++ b/docs/docs/Resources/my-first-pr.md
@@ -7,14 +7,14 @@ It's actually not too hard to do one and it is a great way to contribute. This d
* Double checking that they your edits look good to you on your copy.
* Make a few notes for what you did so people can understand why you made the change.
* Then do a pull request, which is to ask the administrataors of the repository to pull your changes
-back into the dev branch of the main repository.
+back into the dev (not master) branch of the main repository.
* At which time they will do a quick review and merge your changes in.
OK, let's get started. For our example we are going to make an edit to the openaps docs repository.
1. Go to https://github.com/openaps/docs and hit Fork in the upper right to make your own copy of the repository.
2. Github will automatically take you to your copy (notice in the address bar you are now in your own personal github directory)
-3. In the branch drop down on the upper left of the page we are going to select the dev branch, since that is the one we want to edit.
+3. In the branch drop down on the upper left of the page we are going to select the dev branch instead of master, since that is the one we want to edit.
4. Now we need to find the file we want to edit. Click through the directory structure until you find and are looking at the content of the file you want to change.
5. Next, press the pencil icon in the upper right next to the trash can icon.
6. Make edits to the file as necessary.
@@ -22,14 +22,17 @@ OK, let's get started. For our example we are going to make an edit to the opena
8. Commit the changes.
9. Now look and make sure everyting you changed looks like you meant it to (no typos, etc). If any problems, go back and edit again and save again.
-We now have an improved file that we want to be pulled back into the dev repository at https://github.com/openaps/docs
-
-1. Go to https://github.com/openaps/docs
-2. Select dev from the branch drop down on the upper left (since that's where we'd like the changes to go to)
-3. Press the pull request button
-4. Set up the the branches you are targeting. The easiest way of thinking about the branch range is this: the base branch is where you think changes should be applied, the head branch is what you would like to be applied.
-5. So choose so the base fork as openaps/docs and then the base as dev. The head fork is going to be /docs and the base as dev.
-6. It should show the list of changes you made. If not, you did something wrong so stop here and ask for help. If the list looks like your changes then put a note in there to what the overarching reason for the changes are (in your case you only made one, but you could have made a bunch). Create the PR.
+We now have an improved file that we want to be pulled back into the dev branch of the openaps/docs repository at https://github.com/openaps/docs
+
+1. Go to https://github.com/[YOUR_GITHUB_USERNAME]/docs
+ * Or you can go to https://github.com and then click on "docs" in the "Your repositories" section in the lower right. Both methods will get you to the right place.
+2. Select "dev" (not "master") from the branch drop down on the upper left (since that’s where we’d like the changes to go to)
+3. Click the green "New pull request" button
+4. Under the Compare Changes heading, click "compare across forks"
+5. Set up the the branches you are targeting. The easiest way of thinking about the branch range is this: the base branch is where you think changes should be applied, the head branch is what you would like to be applied.
+6. So, choose the base fork as openaps/docs and then the base as dev (not master). The head fork is going to be youraccount/docs and the base as dev (not master).
+
+7. It should show the list of changes you made. If not, you did something wrong so stop here and ask for help. If the list looks like your changes then put a note in there to what the overarching reason for the changes are (in your case you only made one, but you could have made a bunch). Create the PR.
It will now be in a list of PR's that the team will review and potentially give feedback on before committing to the main documentation for openaps!
diff --git a/docs/docs/Resources/troubleshooting.md b/docs/docs/Resources/troubleshooting.md
index f707dfafd..3f648efbb 100644
--- a/docs/docs/Resources/troubleshooting.md
+++ b/docs/docs/Resources/troubleshooting.md
@@ -54,7 +54,7 @@ OpenAPS uses git as the logging mechanism, so it commits report changes on each
To fix a corrupted git repository you can run `oref0-fix-git-corruption.sh`, it will try to fix the repository, and in case when repository is definitly broken it copies the remainings in a safe place (`tmp`) and initializes a new git repo.
-Warning: do not run any openaps commands with sudo in front of it `sudo openaps`. If you do, your .git permissions will get messed up. Sudo should only be used when a command needs root permissions, and openaps does not need that.
+Warning: do not run any openaps commands with sudo in front of it `sudo openaps`. If you do, your .git permissions will get messed up. Sudo should only be used when a command needs root permissions, and openaps does not need that. Such permission problems can be corrected by running `sudo chown -R pi.pi .git` in the openaps directory. If you are using an Intel Edison, run `sudo chown -R edison.users .git`.
### Environment variables
@@ -74,5 +74,7 @@ json: error: input is not JSON: Unexpected '<' at line 1, column 1:
This error usually comes up when you have pulled a file down from Nightscount that was an invalid file. Typcially you might see this when trying to pull down treatments. Make sure that you have your HOST and API_KEY set correctly at the top of your cron, in your ~/.profile
+### Wifi and hotspot issues
+See [wifi troubleshooting page](wifi.md)
diff --git a/docs/docs/walkthrough/phase-0/Can I close the loop with this pump_May 20 2016.jpg b/docs/docs/walkthrough/phase-0/Can I close the loop with this pump_May 20 2016.jpg
new file mode 100644
index 000000000..20152be3d
Binary files /dev/null and b/docs/docs/walkthrough/phase-0/Can I close the loop with this pump_May 20 2016.jpg differ
diff --git a/docs/docs/walkthrough/phase-0/hardware.md b/docs/docs/walkthrough/phase-0/hardware.md
index fcf8c80f7..d52f1de41 100644
--- a/docs/docs/walkthrough/phase-0/hardware.md
+++ b/docs/docs/walkthrough/phase-0/hardware.md
@@ -2,6 +2,10 @@
# Hardware
This section describes the hardware components required for a 'typical' OpenAPS implementation. There are numerous variations and substitutions that can be made but the following items are recommended for getting started. If you come across something that doesn't seem to work, is no longer available, or if you have a notable alternative, feel free to edit this document with your suggestions.
+To start, here is a high-level guide for understanding if your pump is compatible for OpenAPS:
+
+
+
If you're interested in working on communication for another pump (Omnipod, Animas, etc), [click here](http://bit.ly/1nTtccH) to join the collaboration group focusing on alternative pump communication.
@@ -13,7 +17,9 @@ If you're interested in working on communication for another pump (Omnipod, Anim
* 515/715
* 522/722
* 523/723 (with firmware 2.4A or lower)
- * 554 (European Veo, with firmware 2.6A or lower)
+ * 554/754 (European Veo, with firmware 2.6A or lower)
+ * (To check firmware, hit Esc on the home screen and scroll all the way to the bottom. You can also go into the Utilities menu and look for a PC Connect option. If that is present, the pump will *not* work for looping. If it's absent, it should be able to receive temp basal commands.)
+
* A way to communicate with the pump:
* Medtronic CareLink USB stick is the recommended option for your initial loop setup
* **Note** that there are now other hardware options available to communicate with the pump. Some positives to an alternative include better range; some negatives include having to solder and the fact that they're not documented in this set of documentation yet. But if you're interested, check out some of the alternatives in [the mmeowlink wiki](https://github.com/oskarpearson/mmeowlink/wiki).
@@ -121,6 +127,7 @@ A large-capacity power supply that is greater than 8000 mAh (milliAmp-hours) is
[TeckNet® POWER BANK 9000mAh USB External Battery Backup Pack](http://www.amazon.com/gp/product/B00FBD3O2M)
+[Zendure® 2nd Gen A3 Portable Charger 10000mAh - 2.1a Dual USB - in-line charging](www.amazon.com/Zendure-2nd-Portable-Charger-10000mAh/dp/B014RBEAQC/ref=sr_1_1)
### USB Cables
@@ -138,10 +145,9 @@ Repeated wireless communication with the pump drains the battery quite quickly.
### Cases
-The Raspberry Pi is extremely minimalistic and does not come in a protective case. This is fine for development work, but presents an issue for day-to-day use. There are hundreds of cases available, but here some examples of what others are using in their OpenAPS builds.
+The Raspberry Pi is extremely minimalistic and does not come in a protective case. This is fine for development work, but presents an issue for day-to-day use. There are hundreds of cases available, but here is an example of what others are using in their OpenAPS builds.
[JBtek® Jet Black Case for Raspberry Pi B+ & Raspberry Pi 2 Model B](http://www.amazon.com/gp/product/B00ONOKPHC)
-[Raspberry Pi B+ /PI2 Acrylic Case](http://www.amazon.com/Raspberry-Pi-PI2-Acrylic-Case/dp/B00M9ZW6QU)
Additionally, for mobile use, it is helpful to have something besides a lunchbox to carry the entire rig around. The size and weight of the component set as well as the limited range of the CareLink USB stick constrains the options here, but there are still some workable solutions. Waist-worn running gear and camera cases seem to work well. Two options: [FlipBelt](https://flipbelt.com/) and [Lowepro Dashpoint 20](http://store.lowepro.com/dashpoint-20).
diff --git a/docs/docs/walkthrough/phase-0/loops-in-progress.md b/docs/docs/walkthrough/phase-0/loops-in-progress.md
index 9f8d54189..f0bf41d1a 100644
--- a/docs/docs/walkthrough/phase-0/loops-in-progress.md
+++ b/docs/docs/walkthrough/phase-0/loops-in-progress.md
@@ -2,7 +2,9 @@
To get you comfortable with submitting a "PR" (stands for pull request), test it out by submitting a PR to this page, adding your name to the list of people who have loops in progress. This way we know how many people are in the development phase, too.
-New to Github, and PRs? Check out how to submit your first PR.
+New to Github, and PRs? [Check out how to submit your first PR](../../../../docs/docs/Resources/my-first-pr.md).
+
+When you submit the PR, be sure to target the *dev* branch of openaps/docs, not the master branch. If you target master, we'll need to ask you to re-submit, or it will likely cause a merge conflict with edits from people who correctly targeted dev.
List of people who are working on closed loops:
@@ -11,7 +13,16 @@ List of people who are working on closed loops:
- Chris Hannemann
- Sarah Howard
- Mike Stebbins
+- Scott Hanselman
+- Greg Scull
- Aaron Michelson
- Jayson EWER --Intel Edison w/ TI--cc1111
- Frank Best
+- Brooke Armstrong & Matt Pazoles
+- David Young
+- Paul Martin
+- Jarred Yaw
+- Shane Mitchell
- Boris and Kayley Raskin
+- Andy Pabari
+- Rob Kresha - (Papillion, NE, USA)
diff --git a/docs/docs/walkthrough/phase-0/rpi.md b/docs/docs/walkthrough/phase-0/rpi.md
index a5244f9c9..1a1604553 100644
--- a/docs/docs/walkthrough/phase-0/rpi.md
+++ b/docs/docs/walkthrough/phase-0/rpi.md
@@ -98,70 +98,32 @@ You can now skip to [Test SSH Access](#test-ssh-access) and SSH into your RPi2.
### Path 3: Headless WiFi configuration (Windows/Linux only)
Keep the SD card in the reader in your computer. In this step, the WiFi interface is going to be configured in Raspbian, so that we can SSH in to the RPi2 and access the device remotely, such as on a computer or a mobile device via an SSH client, via the WiFi connection that we configure. Go to the directory where your SD card is with all of the files for running Raspbian on your RPi2, and open this file in a text editor.
-`/path/to/sd/card/etc/network/interfaces`
+`/path/to/sd/card/etc/wpa_supplicant/wpa_supplicant.conf`
-Edit the file so it looks like this:
-
-```
-auto lo
-iface lo inet loopback
-iface eth0 inet dhcp
-
-auto wlan0
-allow-hotplug wlan0
-iface wlan0 inet dhcp
-wpa-ssid
-wpa-psk
-```
-
-Replace `` and `` with your own credentials (just text, no quotes). Save the file (without adding any additional extensions to the end of the filename).
-
-Alternatively if you want to connect to known networks automatically when roaming lets say your home wifi and your mobile hotspot you can use the following configuration:
-
-Edit the main network config file and change the WiFi related settings to:
-```
-# the auto wlan0 below is mandatory
-# change wlan0 inet to manual
-# add wpa-roam line
-# define a number of known network using user defined strings, e.g. mobile, home and also default
-
-auto wlan0
-iface wlan0 inet manual
- wpa-roam /etc/wpa_supplicant/wpa_supplicant.conf
-iface mobile inet dhcp
-iface home inet dhcp
-iface default inet dhcp
-```
-Next edit etc/wpa_supplicant/wpa_supplicant.conf and add the following configuration:
+In this file you will list your known WiFi networks so your Pi can connect automatically when roaming (e.g., between your home WiFi and your mobile hotspot).
```
ctrl_interface=DIR=/var/run/wpa_supplicant GROUP=netdev
update_config=1
network={
ssid="YOURMOBILESSID"
- scan_ssid=1
- key_mgmt=WPA-PSK
psk="YOURMOBILEPASS"
- id_str="mobile"
- priority=5
}
network={
ssid="YOURHOMESSID"
- scan_ssid=1
- key_mgmt=WPA-PSK
psk="YOURHOMEPASS"
- id_str="home"
- priority=5
}
```
You can add as many network as you need, the next reboot your system will connect to the first available network listed in your config files. Once the network to which your board is connected becomes unavailable, it start looking for any other known network in the area, and it connects to it if available.
+If you want to connect to a router which doesn't broadcast an SSID, add a line with `scan_ssid=1` after the `ssid` and `psk` lines for that network. (More info and examples for the options you can specify for each network are [here](https://www.freebsd.org/cgi/man.cgi?wpa_supplicant.conf%285%29).)
+
Boot your Pi. (Put the SD card into the RPi2. Plug in the compatible USB WiFi adapter into a RPi2 USB port. Get a micro USB cable and plug the micro USB end into the side of the RPi2 and plug the USB side into the USB power supply.)
If you are unable to access this file on your computer:
* Connect your Pi to your computer with an ethernet cable and boot your Pi
* Log in using PuTTY. The Host Name is `raspberrypi.local` and the Port is 22. The login is `pi` and the password is `raspberry`.
-* Type `sudo nano /etc/network/interfaces` and edit the file as described above, or follow the OS X directions below.
+* Type `sudo nano /etc/wpa_supplicant/wpa_supplicant.conf` and edit the file as described above.
## Test SSH Access
@@ -205,11 +167,11 @@ Run
`sudo raspi-config`
-to expand filesystem, change user password and set timezone (in internationalization options). This will take effect on the next reboot, so go ahead and reboot if prompted, or run `sudo reboot` when you're ready.
+Here you can expand filesystem to maximize memory, change user password and set timezone (in internationalization options). This will take effect on the next reboot, so go ahead and reboot if prompted, or run `sudo reboot` when you're ready.
-## Setup Password-less Login [optional]
+## Setting up an SSH key for Password-less Login [optional]
-We will now setup a public/private key identity, and configure your local computer and the Raspberry Pi to automatically use it, in order to allow convenient future ssh access to the Pi without requiring a password.
+You can setup a public/private key identity, and configure your local computer and the Raspberry Pi to automatically use it. This will allow SSH access to the Pi without requiring a password. Some people find this feature very convenient.
### Windows
@@ -241,12 +203,22 @@ Instead of appending it to the list of authorized keys, you may simply copy your
Finally, `ssh pi@raspberrypi.local` to make sure you can log in without a password.
-### Disabling password login [optional]
-To secure the Pi, you should either set a password (using `sudo raspi-config` above, or with `sudo passwd`), or disable password login completely. If you want to disable password login (so you can only log in with your ssh key), open the `sshd_config` file in nano text editor on the Pi as follows
+### Disabling password login [optional - WARNING: THIS COULD POTENTIALLY LOCK YOU OUT OF YOUR RASPBERRY PI]
+Make sure you fully understand this feature before proceeding. It is completely optional. There are two ways of securing the Pi:
+
+1) Setting a password:
+
+ a) use `sudo raspi-config` as described above, or
+
+ b) `sudo passwd`
+
+2) Disabling password login completely. In this case, you can ONLY log in with your SSH key. Be careful here.
+
+ a) Open the `sshd_config` file in nano text editor on the Pi as follows
`sudo nano /etc/ssh/sshd_config`
-Change the following
+ b) Change the following
```
PermitRootLogin yes
@@ -262,7 +234,7 @@ PasswordAuthentication no
Note that the second line was previously commented out.
-From now on you will be able to SSH in with your private SSH key only.
+From now on you will be able to SSH in with your private SSH key ONLY.
## Wifi reliability tweaks [optional]
@@ -310,3 +282,92 @@ Update the RPi2.
`sudo apt-get update && sudo apt-get -y upgrade`
The packages will take some time to install.
+
+## Configure Bluetooth Low Energy tethering [optional]
+
+The Raspberry Pi can be tethered to a smartphone and share the phone's internet connection. Bluetooth tethering needs to be enabled and configured on the phone device and your carrier/plan must allow tethering. The Raspberry Pi 3 has an inbuilt Bluetooth Low Energy (BLE) chip, while a BLE USB dongle can be used with the other Pi models.
+
+The main advantages of using BLE tethering are that it consumes less power on the phone device than running a portable WiFi hotspot and it allows the Raspberry Pi to use whatever data connection is available on the phone at any given time - e.g. 3G/4G or WiFi. Some have also found that power consumption on the Raspberry Pi is lower when using BLE tethering compared to using a WiFi connection, although this may vary depending on BLE USB dongle, WiFi dongle, etc.
+
+First, we clone a repository which contains scripts which are used later in the setup -
+
+```
+cd /home/pi
+git clone https://github.com/WayneKeenan/RaspberryPi_BTPAN_AutoConnect.git
+```
+
+We then copy the required scripts into a 'bin' directory -
+```
+mkdir -p /home/pi/bin
+cp /home/pi/RaspberryPi_BTPAN_AutoConnect/bt-pan /home/pi/bin
+cp /home/pi/RaspberryPi_BTPAN_AutoConnect/check-and-connect-bt-pan.sh /home/pi/bin
+```
+
+To configure a connection from the command line -
+
+`sudo bluetoothctl`
+
+Enter the following commands to bring up the adapter and make it discoverable -
+
+```
+power on
+discoverable on
+agent on
+default-agent
+```
+
+The adapter is now discoverable for three minutes. Search for bluetooth devices on your phone and initiate pairing. The process varies depending on the phone and the dongle in use. The phone may provide a random PIN and bluetoothctl may ask you to confirm it. Enter 'yes'. Then click 'pair' on the phone. Instead, the phone may ask you to enter a PIN. If so, enter '0000' and when bluetoothctl asks for a PIN, enter the same code again. Either way, bluetoothctl should inform you that pairing was successful. It will then ask you to authorize the connection - enter 'yes'.
+
+Execute the paired-devices command to list the paired devices -
+
+```
+paired-devices
+Device AA:BB:CC:DD:EE:FF Nexus 6P
+```
+
+Your paired phone should be listed (in this example, a Google Nexus 6P). Copy the bluetooth address listed for it; we will need to provide this later.
+
+Now trust the mobile device (notice that bluetoothctl features auto-complete, so you can type the first few characters of the device's bluetooth address (which we copied previously) and hit to complete the address.
+
+NOTE: Whenever you see 'AA:BB:CC:DD:EE:FF' or 'AA_BB_CC_DD_EE_FF' in this guide, replace it with the actual address of your mobile Bluetooth device, in the proper format (colons or underscores).
+
+`trust AA:BB:CC:DD:EE:FF`
+
+Quit bluetoothctl with 'quit'.
+
+Now, we create a service so that a connection is established at startup. Execute the following commands to create a net-bnep-client.service file and open it for editing in Nano -
+
+```
+cd /etc/systemd/system
+sudo nano net-bnep-client.service
+```
+
+In the editor, populate the file with the text below, replacing AA:BB:CC:DD:EE:FF with the address noted earlier -
+
+```
+[Unit]
+After=bluetooth.service
+PartOf=bluetooth.service
+
+[Service]
+ExecStart=/home/pi/bin/bt-pan client AA:BB:CC:DD:EE:FF
+
+[Install]
+WantedBy=bluetooth.target
+```
+
+Save the file, then enable the service -
+
+`sudo systemctl enable net-bnep-client.service`
+
+Open your crontab for editing -
+
+`crontab -e`
+
+...and add an entry to check the connection every minute and reconnect if necessary -
+
+`* * * * * /home/pi/bin/check-and-connect-bt-pan.sh`
+
+Save the file, then restart -
+
+`sudo shutdown -r now`
diff --git a/docs/docs/walkthrough/phase-1/visualization.md b/docs/docs/walkthrough/phase-1/visualization.md
index 54371d668..b0240dbc2 100644
--- a/docs/docs/walkthrough/phase-1/visualization.md
+++ b/docs/docs/walkthrough/phase-1/visualization.md
@@ -8,18 +8,17 @@ The integration requires setting up Nightscout and making changes and additions
### Nightscout Setup
-OpenAPS requires the latest (currently dev) version of Nighthscout, which can be found here: https://github.com/nightscout/cgm-remote-monitor/tree/dev.
+OpenAPS requires the latest (currently dev) version of Nightscout, which can be found here: https://github.com/nightscout/cgm-remote-monitor/tree/dev.
-Note: currently there is a bug in the dev version, which doesn't allow you to set up a new profile using the profile editor. If you are starting a fresh install of Nightscout, you should first deploy the master version of the code. Once the master version is up an running, you can create your profile with information on basal rates, etc. After that, you can deploy the dev version. If you have an existing version of Nightscout, then make sure you create your profile before moving to the dev version. Or, you may keep your existing Nightscout as is, and start a new Nightscout deployment (master first, followed by dev), specifically to test OpenAPS integration.
-The steps discussed here are essantially the same for both Azure and Heroku users. Two configuration changes must be made to the Nightscout implementation:
+The steps discussed here are essentially the same for both Azure and Heroku users. Two configuration changes must be made to the Nightscout implementation:
* Add "openaps" (without the quotes) and, optionally, "pump" (without the quotes) to the list of plugins enabled, and
* Add a new configuration variable DEVICESTATUS_ADVANCED="true" (without the quotes)
For Azure users, here is what these configuration changes will look like (with just "openaps" added): 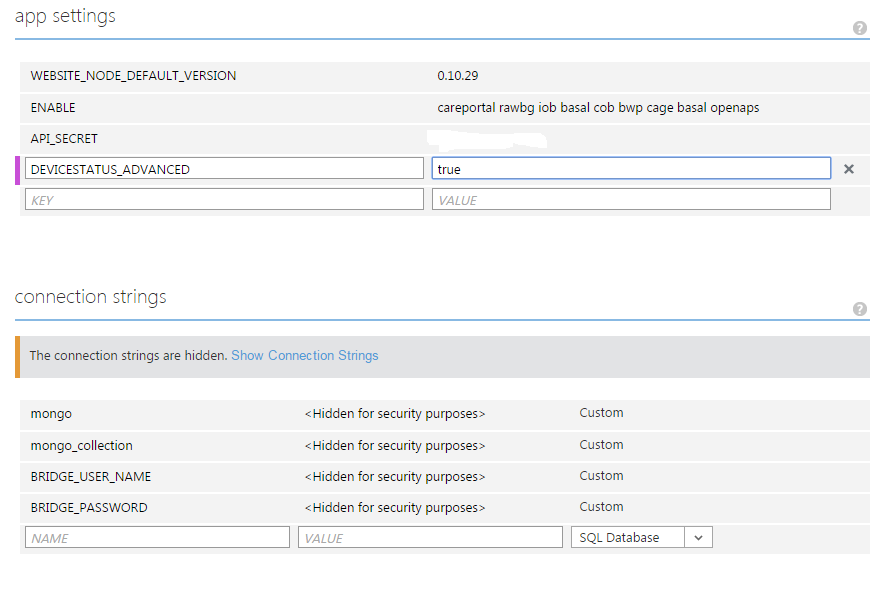. For Heroku users, exactly the same changes should be made on the Config Vars page. The optional "pump" plugin enables additional pump monitoring pill boxes. For example, assuming you have added "pump" to the list of enabled plugins, you may add a new configuration variable PUMP_FIELDS="reservoir battery" to display pump reservoir and battery status on the Nightscout page. The "pump" plugin offers a number of other options, as documented on the Nightscout readme page: https://github.com/nightscout/cgm-remote-monitor/blob/dev/README.md#built-inexample-plugins
-Next, on your Nightscout website, go to the Settings (3 horizontal bars) in the upper right corner. At the very bottom of the Settings menu, in the "About" section, you may check the Nightscout version (e.g. version 0.9.0-dev). Just above is a list of Plugins available. OpenAPS should show up. Click the check box to enable. Similarly, in the case you've enabled the "pump" plugin, "Pump" should also show up in the list, and you may chekc the box to enable. You should now see the OpenAPS pill box (and any optional pump monitoring pill boxes) on the left side of the Nightscout page near the time. You may also want to graphically show the basal rates: select "Default" or "Icicle" from the "Render Basal" pull-down menu in the Settings.
+Next, on your Nightscout website, go to the Settings (3 horizontal bars) in the upper right corner. At the very bottom of the Settings menu, in the "About" section, you may check the Nightscout version (e.g. version 0.9.0-dev). Just above is a list of Plugins available. OpenAPS should show up. Click the check box to enable. Similarly, in the case you've enabled the "pump" plugin, "Pump" should also show up in the list, and you may check the box to enable. You should now see the OpenAPS pill box (and any optional pump monitoring pill boxes) on the left side of the Nightscout page near the time. You may also want to graphically show the basal rates: select "Default" or "Icicle" from the "Render Basal" pull-down menu in the Settings.
### Environment Variables for OpenAPS Access to Nightscout
@@ -37,14 +36,25 @@ API_SECRET=; export API_SECRET
Now run ```source /etc/profile``` to enact the changes we've just made without restart the machine
+### Configure Nightscout profile
+
+You need to create a profile in your Nightscout site that contains the Timezone, Duration of Insulin Activity (DIA), Insulin to carb ratio (I:C), Insulin Sensitivity Factor (ISF), Carbs Activity / Absorption rate, Basal Rates and Target BG range.
+
+These settings are not currently updated from the values stored in the pump. You will need to keep the Nightscout profile in sync with any changes you make in your pump.
+
+To configure your profile, on your Nightscout website, go to the Settings (3 horizontal bars) in the upper right corner.
+Click on the Profile Editor button.
+Create a new profile (if you don't already have one) using the settings that match what you already have set up in your pump.
+Fill out all the profile fields and click save.
+
### Configuring and Uploading OpenAPS Status
Integration with Nightscout requires couple of changes to your OpenAPS implementation, which include:
* Adding a new `ns-status` device, and generating a new report `monitor/upload-status.json`, which consolidates the current OpenAPS status to be uploaded to Nightscout
-* Uploading the status report to Nightscount, using the `ns-upload` command
+* Uploading the status report to Nightscout, using the `ns-upload` command
-Upon successful completion of these two steps, you will be able to see the current OpenAPS status by hovering over the OpenAPS pill box on your Nightscount page, as shown here, for example: 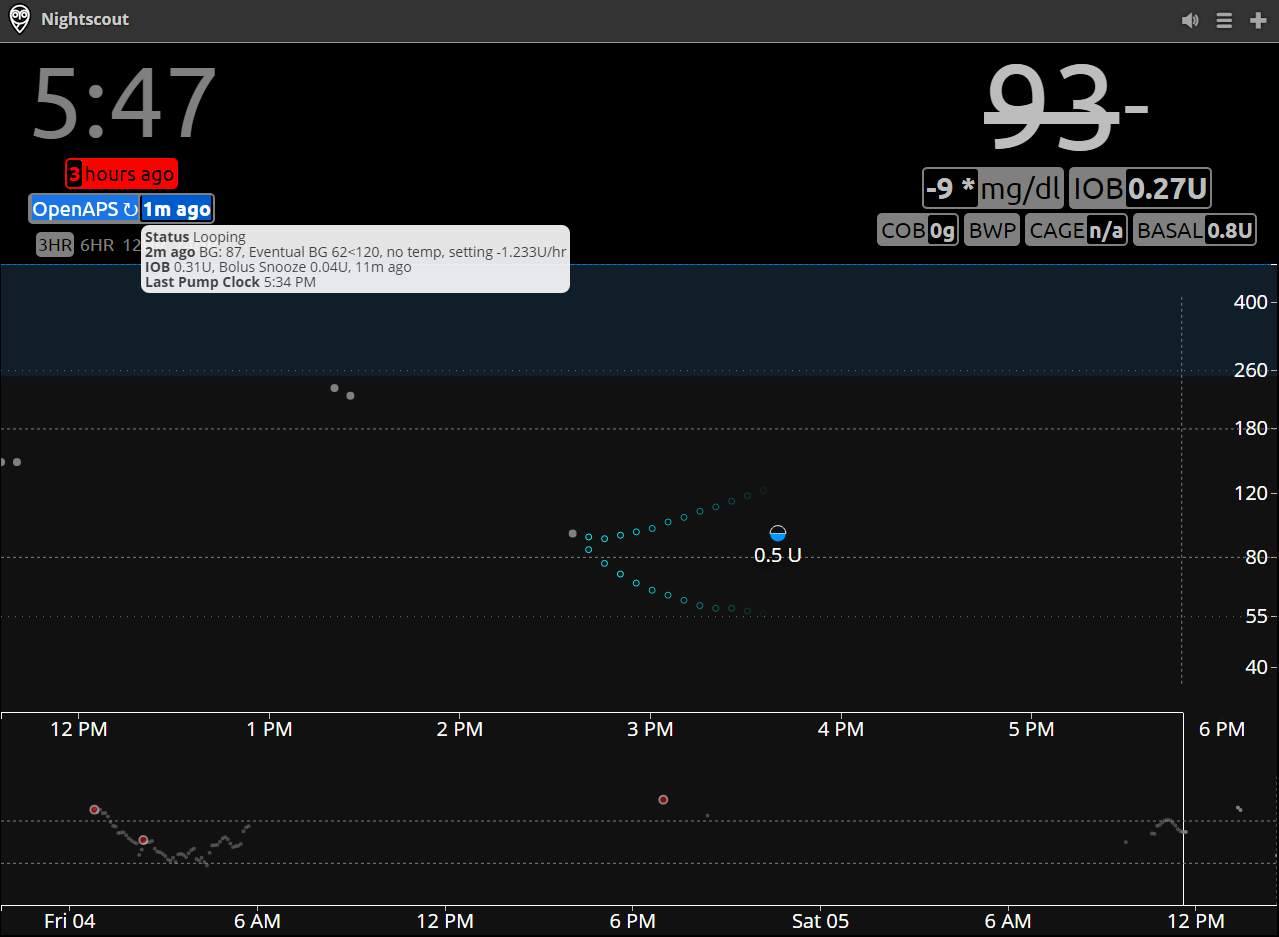
+Upon successful completion of these two steps, you will be able to see the current OpenAPS status by hovering over the OpenAPS pill box on your Nightscout page, as shown here, for example: 
The `ns-status` is a virtual device in the oref0 system, which consolidates OpenAPS status info in a form suitable for upload to Nightscout. First, add the device:
@@ -69,13 +79,14 @@ $ openaps report add monitor/upload-status.json JSON ns-status shell monitor/clo
The reports required to generate upload-status.json should look familiar. If you have not generated any of these required reports, you should set them up and make sure they all work. In particular, note that monitor/clock-zoned.json contains the current pump clock time stamp, but with the timezone info included. If you have not generated that report already, you may do so using the following commands, which add a `tz` virtual device and use it to create clock-zoned.json starting from clock.json.
```
+$ sudo pip install recurrent
$ openaps vendor add openapscontrib.timezones
$ openaps device add tz timezones
$ git add tz.ini
$ openaps report add monitor/clock-zoned.json JSON tz clock monitor/clock.json
```
-At this point, you may want to update your monitor-pump alias to make sure that it produces all the required reports, so that uploading status to Nightscount can be automated. After you've generated a monitor/upload-status.json report, you can try to manually upload the OpenAPS status to Nightscout using the `ns-upload` command:
+At this point, you may want to update your monitor-pump alias to make sure that it produces all the required reports, so that uploading status to Nightscout can be automated. After you've generated a monitor/upload-status.json report, you can try to manually upload the OpenAPS status to Nightscout using the `ns-upload` command:
```
$ ns-upload $NIGHTSCOUT_HOST $API_SECRET devicestatus.json monitor/upload-status.json
@@ -114,7 +125,9 @@ $ openaps alias add format-latest-nightscout-treatments '! bash -c "nightscout c
$ openaps alias add upload-recent-treatments '! bash -c "openaps format-latest-nightscout-treatments && test $(json -f upload/latest-treatments.json -a created_at eventType | wc -l ) -gt 0 && (ns-upload $NIGHTSCOUT_HOST $API_SECRET treatments.json upload/latest-treatments.json ) || echo \"No recent treatments to upload\""' || die "Can't add upload-recent-treatments"
```
-Note that a pumphistory-zoned.json report is required, which can be generated from pumphistory.json using `tz`, following the approach described above for clock-zoned.json. After running your loop from command line, you may try executing `openaps upload-recent-treatments` manually from command line. Upon successful upload, the recent treatments will show up automatically on the Nightscount page.
+Note that a pumphistory-zoned.json report is required, which can be generated from pumphistory.json using `tz`, following the approach described above for clock-zoned.json, including making sure to add it to your monitor-pump alias. In addition, if you haven't already created a settings/model.json report, you should create that report and invoke it since it is required for format-latest-nightscout-treatments.
+
+After running your loop from command line, you may try executing `openaps upload-recent-treatments` manually from command line. Upon successful upload, the recent treatments will show up automatically on the Nightscount page.
Note: Currently extended boluses are not handled well and depending on the timing of the upload are either missed entirely or have incorrect information.
diff --git a/docs/docs/walkthrough/phase-2/Using-oref0-tools.md b/docs/docs/walkthrough/phase-2/Using-oref0-tools.md
index 7fb577310..5f4f55536 100644
--- a/docs/docs/walkthrough/phase-2/Using-oref0-tools.md
+++ b/docs/docs/walkthrough/phase-2/Using-oref0-tools.md
@@ -12,7 +12,7 @@ $ openaps device add oref0 process oref0
and then you can be more specific and add individual oref0 processes as virtual devices using the following commands:
```
-$ openaps device add get-profile process --require "settings bg_targets insulin_sensitivities basal_profile max_iob" oref0 get-profile
+$ openaps device add get-profile process --require "settings bg_targets insulin_sensitivities basal_profile preferences" oref0 get-profile
$ openaps device add calculate-iob process --require "pumphistory profile clock" oref0 calculate-iob
$ openaps device add determine-basal process --require "iob temp_basal glucose profile" oref0 determine-basal
```
@@ -42,34 +42,52 @@ The purpose of the `get-profile` process is to consolidate information from mult
```
$ openaps report add settings/bg_targets_raw.json JSON pump read_bg_targets
```
-To convert this "raw" file
-The units function assures that units will match
-add a report that will perform
-$ openaps use units bg_targets settings/bg_targets_raw.json
-and output not to the screen but into a file called settings/bg_targets.json
-add units to your list of devices with
-$ openaps device add units units
+If your pump OR CGM is European and displays mmol/L as opposed to mg/dl you will need to convert this "raw" file.
+
+First install the unit conversion device to ensure all units will match.
+ ```
+ $ openaps device add units units
+ ```
+Go through the standard process of use, report add, report invoke for the 2 reports below.
+
+For Blood Sugar Conversion
+The `units` function ensures that units will match. To use it, add units to your list of devices with:
+ ```
+ $ openaps device add units units
+ ```
+To convert this "raw" file, we need to add a report that will perform
+`$ openaps use units bg_targets settings/bg_targets_raw.json`
+and output not to the screen but into a file called settings/bg_targets.json:
+ ```
+ $ openaps report add settings/bg_targets.json JSON units bg_targets settings/bg_targets_raw.json
+ ```
+For Insulin Sensitivity
+ ```
+ $ openaps report add settings/insulin_sensitivities.json JSON units insulin_sensitivities_raw.json
+ ```
* `insulin_sensitivities` outputs a JSON file with insulin sensitivites obtained from the pump:
```
- $ openaps report add settings/insulin_sensitivities_raw.json JSON pump read_insulin_sensitivies
+ $ openaps report add settings/insulin_sensitivities_raw.json JSON read_insulin_sensitivies
```
-To convert this "raw" file you must do the same as done for bg_targets_raw
-to obtain the file settings/insulin_sensitivities
* `basal_profile` outputs a JSON file with the basal rates stored on the pump in your basal profile
```
- $ openaps report add settings/basal_profile.json JSON pump read_basal_profile_std
+ $ openaps report add settings/basal_profile.json JSON read_basal_profile_std
```
-* `max_iob` is an exception: in contrast to the other settings above, `max_iob` is not the result of an openaps report. It's a JSON file that should contain a single line, such as: `{"max_iob": 2}`. You can create this file by hand, or use the [oref0-mint-max-iob](https://github.com/openaps/oref0/blob/master/bin/oref0-mint-max-iob.sh) tool to generate the file. The max_iob variable represents an upper limit to how much insulin on board oref0 is allowed to contribute by enacting temp basals over a period of time. In the example above, `max_iob` equals 2 units of insulin.
+* `preferences` is an exception: in contrast to the other settings above, `preferences` is not the result of an openaps report. It's a JSON file that should contain a single line with your maximum IOB, such as: `{"max_iob": 2}`. You can create this file by hand, or use the [oref0-mint-max-iob](https://github.com/openaps/oref0/blob/master/bin/oref0-mint-max-iob.sh) tool to generate the file. The `max_iob` variable represents an upper limit to how much insulin on board oref0 is allowed to contribute by enacting temp basals over a period of time. In the example above, `max_iob` equals 2 units of insulin.
Make sure you test invoking each of these reports as you set them up, and review the corresponding JSON files using `cat`. Once you have a report for each argument required by `get-profile`, you can add a `profile` report:
```
-$ openaps report add settings/profile.json text get-profile shell settings/settings.json settings/bg_targets_raw.json settings/bg_targets.json settings/insulin_sensitivities_raw.json settings/insulin_sensitivities.json settings/basal_profile.json max_iob.json
+$ openaps report add settings/profile.json text get-profile shell settings/settings.json settings/bg_targets_raw.json settings/insulin_sensitivities_raw.json settings/basal_profile.json preferences.json
+```
+If you need to add a `profile` report that has the unit conversion, you can use this:
+```
+$ openaps report add settings/profile.json text get-profile shell settings/settings.json settings/bg_targets.json settings/insulin_sensitivities.json settings/basal_profile.json preferences.json
```
Note how the `profile` report uses `get-profile` virtual device, with all the required inputs provided.
diff --git a/docs/docs/walkthrough/phase-2/building-preflight-safety-checks.md b/docs/docs/walkthrough/phase-2/building-preflight-safety-checks.md
index 5f43bbd1e..9ef27c200 100644
--- a/docs/docs/walkthrough/phase-2/building-preflight-safety-checks.md
+++ b/docs/docs/walkthrough/phase-2/building-preflight-safety-checks.md
@@ -4,7 +4,7 @@ Before moving on to consolidating all of these capabilities into a single alias,
* There are several potential issues that may adveresely affect operation of the system. For example, RF communication with the pump may be compromised. It has also been observed that the CareLink USB stick may become unresponsive or "dead", requiring a reset of the USB ports. Furthermore, in general, the system should not act on stale data. Let's look at some approaches you may consider to address these issues.
-Ensuring that your openaps implementation can't act on stale data could be done by deleting all of the report files in the `monitor` directory before the reports are refreshed. YOu may simply use `rm -f` bash command, which removes file(s), while ignoring cases when the file(s) do not exist. If a refresh fails, the data required for subsequent commands will be missing, and they will fail to run. For example, here is an alias that runs the required bash commands:
+Ensuring that your openaps implementation can't act on stale data could be done by deleting all of the report files in the `monitor` directory before the reports are refreshed. You may simply use `rm -f` bash command, which removes file(s), while ignoring cases when the file(s) do not exist. If a refresh fails, the data required for subsequent commands will be missing, and they will fail to run. For example, here is an alias that runs the required bash commands:
```
openaps alias add gather '! bash -c "rm -f monitor/*; openaps gather-profile && openaps monitor-cgm && openaps monitor-pump && openaps report invoke monitor/iob.json"'
@@ -37,7 +37,7 @@ returns the current pump time stamp, such as "2016-01-09T10:47:56", if the syste
Collecting all the error checking, a `preflight` alias could be defined as follows:
```
-$ openaps alias add preflight '! bash -c "rm -f monitor/clock.json && openaps report invoke monitor/clock.json 2>/dev/null && grep -q T monitor/clock.json && echo PREFLIGHT OK || (mm-stick warmup || (sudo oref0-reset-usb && echo PREFLIGHT SLEEP && sleep 120); echo PREFLIGHT FAIL; exit 1)"'
+$ openaps alias add preflight '! bash -c "rm -f monitor/clock.json && openaps report invoke monitor/clock.json 2>/dev/null && grep -q T monitor/clock.json && echo PREFLIGHT OK || (mm-stick warmup || sudo oref0-reset-usb; echo PREFLIGHT FAIL; sleep 120; exit 1)"'
```
In this `preflight` example, a wait period of 120 seconds is added using `sleep` bash command if the USB ports have been reset in an attempt to revive the MM CareLink stick. This `preflight` example also shows how bash commands can be chained together with the bash && ("and") or || ("or") operators to execute different subsequent commands depending on the output code of a previous command (interpreted as "true" or "false").
diff --git a/docs/docs/walkthrough/phase-2/using-openaps-tools.md b/docs/docs/walkthrough/phase-2/using-openaps-tools.md
index 45d25f7c0..15fa9ae8e 100644
--- a/docs/docs/walkthrough/phase-2/using-openaps-tools.md
+++ b/docs/docs/walkthrough/phase-2/using-openaps-tools.md
@@ -169,7 +169,11 @@ Go ahead and try some more pump uses to find out what they do. Note that some of
Now let's try communicating with the Dexcom receiver.
-Hint: Your Dexcom should be nearly fully charged before plugging it in to your Raspberry Pi. If, when you plug in your Dexcom, it causes your WiFi dongle to stop blinking, that is a sign that it is drawing too much power and needs to be charged.
+Hint: Your Dexcom should be nearly fully charged before plugging it in to your Raspberry Pi. If, when you plug in your Dexcom, it causes your WiFi dongle to stop blinking, or if the charging icon on the Dexcom keeps cycling on and off, that is a sign that it is drawing too much power and needs to be charged.
+
+> Workaround: If you continue to have problems, try increasing the mA output to the USB ports, you can do this by running the following command `$ sudo bash -c "echo -e \"#Enable Max USB power\nmax_usb_current=1\" >> /boot/config.txt"`.
+
+> Reboot via `$ sudo shutdown -r now` to pick up the changes.
`$ openaps use iter_glucose 1`
@@ -212,20 +216,20 @@ Some people have found it more beneficial to pull blood glucose values from Nigh
1) Similar like above, we need to create a device that talks to Nightscout. Add this device called "curl" to your list of devices in your openaps.ini file:
```
-[device "curl"]
-fields =
-cmd = bash
-vendor = openaps.vendors.process
-args = -c "curl -s https://yourwebsite.azurewebsites.net/api/v1/entries.json | json -e 'this.glucose = this.sgv'"
+[device "curl"]
+fields =
+cmd = bash
+vendor = openaps.vendors.process
+args = -c "curl -s https://yourwebsite.azurewebsites.net/api/v1/entries.json | json -e 'this.glucose = this.sgv'"
```
In addition, you need to alter your monitor/glucose.json report to use this device rather than the cgms device you setup above. The report will look like this in your openaps.ini file:
```
-[report "monitor/glucose.json"]
-device = curl
-use = shell
-reporter = text
+[report "monitor/glucose.json"]
+device = curl
+use = shell
+reporter = text
```
Many people will actually setup both ways to pull the blood glucose level and switch between the different devices depending on their needs. If you are going to pull it directly from Nightscout then you will have to have internet access for the Raspberry Pi.
@@ -367,3 +371,4 @@ directory will be saved in your online Bitbucket repository. Whenever you would
like to update your backup, simply go into your `` directory and `$
git push`. This process can be automated, but we'll save that for another day.
+[Github mkimg tool](https://github.com/zymbit/rpi-mkimg) has the ability to create a full SD card image with the advantage to shrink it to its minimum size quite different from the windows Win32DiskImager which always creates an image which is as large as the card. Using Win32DiskImager it might not be possible to restore an image on a new card with the same size. Using Github mkimg tool instead for example a 32 GB size card can be reduced to approximately 1.9 GB.
diff --git a/docs/docs/walkthrough/phase-4/troubleshooting-loop.md b/docs/docs/walkthrough/phase-4/troubleshooting-loop.md
index 64612b9d9..350982671 100644
--- a/docs/docs/walkthrough/phase-4/troubleshooting-loop.md
+++ b/docs/docs/walkthrough/phase-4/troubleshooting-loop.md
@@ -52,7 +52,7 @@ If you'd like to help out with defining all the desired behaviors in the form of
2) Type `sudo npm install -g mocha` and `sudo npm install -g should`
-3) You should then be able to run `make` from the oref0 directory to run all of the existing unit tests, or something like `mocha -c tests/determine-basal.test.js 2>&1 | less -r` to run just the determine-basal tests.
+3) You should then be able to run `make` from the ~/src/oref0 directory to run all of the existing unit tests, or something like `mocha -c tests/determine-basal.test.js 2>&1 | less -r` to run just the determine-basal tests.
4) As you add additional unit tests, you'll want to run `make` again after each one.
diff --git a/docs/docs/walkthrough/phase-6/Configure-Automatic-Sensitivity-Mode.md b/docs/docs/walkthrough/phase-6/Configure-Automatic-Sensitivity-Mode.md
index 44184b2a3..0b4ac03e2 100644
--- a/docs/docs/walkthrough/phase-6/Configure-Automatic-Sensitivity-Mode.md
+++ b/docs/docs/walkthrough/phase-6/Configure-Automatic-Sensitivity-Mode.md
@@ -1,30 +1,25 @@
-## Configuring Automatic Sensitivity Mode
+## Configuring Automatic Sensitivity and Meal Assist Mode
For more information review https://github.com/openaps/oref0/issues/58
-1) Ensure to get the latest dev branch
+1) Install the latest dev branch of `oref0`:
```
-cd
-cd src
-cd oref0
-git pull
-git checkout dev
-sudo npm install -g
+sudo npm install -g git://github.com/openaps/oref0.git'#dev'
```
-Make sure you run the last line to install the tools
-2) Next in order to properly execute the new auto-sensitivity module, you need to have at least 24 hour worth of pump history data and enough bg readings (past 24 hours).
-In your openaps.ini apply the following changes:
+2) Next in order for the new auto-sensitivity report to run, you need to have at least 24 hours worth of pump history data and enough bg readings (24 hours).
+In your `openaps.ini` apply the following changes:
```
-[report "[Your Path]/glucose.json"]
+[report "monitor/glucose.json"]
device = cgm
use = iter_glucose
reporter = JSON
count = 288
```
-Note: If using Nightscout add count=288 to your entries.json API call as a querystring parameter
-For NS it will look something like this depending on how you implement it (note the ?count=288) that is what you have to add
+(NOTE: if using Nightscout, add `count=288` to your `entries.json` API call as a querystring parameter)
+
+One way to do this is to go to your openaps directory and edit the `ns-glucose.ini` file. Depending on how you've implemented it, it should look something like this (please note the `?count=288`):
```
[device "curl"]
fields =
@@ -32,17 +27,22 @@ cmd = bash
vendor = openaps.vendors.process
args = -c "curl -s https://[Your URL]/api/v1/entries.json?count=288 | json -e 'this.glucose = this.sgv'"
```
+If your `glucose.json` does not have enough entries you will see a warning when running your `auto-sens.json` report:
+```
+Error: not enough glucose data to calculate autosens.
+```
+3) After applying the above change you need to add a new `auto-sens` device and an `auto-sens` report. Run this command to create the `auto-sens` report:
-If your [glucose.json] does not have enough entries you will see a warning when running your auto-sens.json report "Error: not enough glucose data to calculate autosens."
+`openaps device add auto-sens process --require "glucose pumphistory insulin_sensitivities basal_profile profile" oref0 detect-sensitivity`
-3) After applying the above change you need to add a new device and report as follow:
+If the command exectuted properly, the contents of `openaps.ini` should contain:
```
[device "auto-sens"]
vendor = openaps.vendors.process
extra = auto-sens.ini
```
-and your auto-sens.ini should look like this:
+Another new file named `auto-sens.ini` should have been created, and it should contain:
```
[device "auto-sens"]
fields = glucose pumphistory insulin_sensitivities basal_profile profile
@@ -50,53 +50,58 @@ cmd = oref0
args = detect-sensitivity
```
-4) Next create this report
+4) In order for `auto-sens` to run properly, you need to make sure you pull enough history from your pump - 24 hours plus however many yours you have set for your DIA. To do this, you will create a new report called `pumphistory-24h.json`:
```
-[report "[Your Path]/auto-sens.json"]
-profile = [Your Path]/profile.json
-use = shell
-reporter = text
-json_default = True
-pumphistory = [Your Path]/pump-history-zoned.json
-basal_profile = [Your Path]/active-basal-profile.json
-insulin_sensitivities = [Your Path]/insulin-sensitivities.json
-glucose = [Your Path]/ns-glucose.json
-device = auto-sens
-remainder = []
+openaps report add settings/pumphistory-24h.json JSON pump iter_pump_hours 28
```
+(NOTE: the `28` assumes a 4h DIA - please adjust accordingly)
-5) Next we need to pass auto-sens.json to oref0-determine-basal.json, in openaps.ini add a new input simillar to folowing example below:
+5) Once the device is created, we need to create the `auto-sens.json` report. Run this command to create the `auto-sens.json` report:
+```
+openaps report add settings/auto-sens.json text auto-sens shell monitor/glucose.json settings/pumphistory-24h.json settings/insulin_sensitivities.json settings/basal_profile.json settings/profile.json []
+```
+Now invoke the report to test:
+```
+openaps invoke report settings/auto-sens.json
+```
+
+6) Next we need to add the `auto-sens.json` report to the `oref0-determine-basal` device. In `openaps.ini` make sure your `oref0-determine-basal` looks similar to this:
```
[device "oref0-determine-basal"]
-fields = iob current-temps glucose profile auto-sens meal
+fields = iob current-temps glucose profile **auto-sens** meal
cmd = oref0-determine-basal
vendor = openaps.vendors.process
args =
+```
+(NOTE: in the `fields` above, `meal` should only be present if meal assist is configured)
+7) At this point, in the process you should already have an `enact/suggested.json` report. Edit your `openaps.ini` file and add the bottom line to that report:
+```
[report "enact/suggested.json"]
-profile = [Your Path]/profile.json
+profile = settings/profile.json
use = shell
reporter = text
-current-temps = [Your Path]/temp-basal-status.json
+current-temps = monitor/temp-basal-status.json
device = oref0-determine-basal
-iob = [Your Path]/iob.json
-glucose = [Your Path]/glucose.json
-meal = [Your Path]/meal.json
-auto-sens = [Your Path]/auto-sens.json
+iob = monitor/iob.json
+glucose = monitor/glucose.json
+meal = monitor/meal.json
+auto-sens = settings/auto-sens.json
```
+(NOTE: as stated above, if you do not have meal assist enabled, do not include the `meal` line)
-6) Next you have to make sure you pull 28 hours (24h + DIA) of pump history. Change the hours to 24, so your pump history report should look like this:
+8) Based on the configuration of the basic loop, it is recommended that the `settings/auto-sens.json` be added to the `gather-profile` alias:
```
-[report "[Your Path]/pump-history.json"]
-device = pump
-hours = 28
-use = iter_pump_hours
-reporter = JSON
+gather-profile report invoke settings/settings.json settings/bg_targets.json settings/insulin_sensitivities.json settings/basal_profile.json settings/profile.json monitor/auto-sens.json
```
-Note. Your loop should run without auto-sens.json report but if you don't pass that as an input you will see the following message while executing oref0-determine-basal.js:
+and that the `gather` alias be adjusted to make sure `gather-profile` is at the end. This is because the `settings/auto-sens.json` report depends upon elements from the preceding two aliases to run.
-Optional feature Auto Sensitivity not enabled: { [Error: ENOENT: no such file or directory, open 'online'] errno: -2, code: 'ENOENT', syscall: 'open', path: 'online' }
+`gather ! bash -c "rm -f monitor/*; openaps monitor-cgm && openaps monitor-pump && openaps gather-profile"`
+Note. Your loop should run without `auto-sens.json` report but if you don't pass that as an input you will see the following message while executing `oref0-determine-basal.js`:
+```
+Optional feature Auto Sensitivity not enabled: { [Error: ENOENT: no such file or directory, open 'online'] errno: -2, code: 'ENOENT', syscall: 'open', path: 'online' }
+```
Here is an example of running the loop with Auto Sensitivity feature enabled:
```
Feb 26 22:26:11 raspberrypi openaps-loop: reporting oref0-prepare/mm-normalized.json
@@ -152,7 +157,3 @@ Feb 26 22:26:42 raspberrypi openaps-loop: "reason": "Eventual BG 104>100 but A
Feb 26 22:26:42 raspberrypi openaps-loop: }
Feb 26 22:26:43 raspberrypi openaps-loop: No recommendation to send
```
-
-
-
-
diff --git a/docs/docs/walkthrough/phase-6/Usability-considerations.md b/docs/docs/walkthrough/phase-6/Usability-considerations.md
new file mode 100644
index 000000000..b40cd9269
--- /dev/null
+++ b/docs/docs/walkthrough/phase-6/Usability-considerations.md
@@ -0,0 +1,19 @@
+#Usability Considerations
+
+Now that you've closed the loop, you probably have a lot of new "first" experiences to deal with. Like much of this looping experience, you'll figure it out as you go along, and figure out what's right for you. But here are some ideas or tips to consider:
+
+* **What do you do with the loop in airport security when you travel**
+
The loop is off the shelf hardware - it's no different than your phone or other small gadgets, so leave it in your carry-on bag when going through security. (Dana note: I have traveled well over 100 times with my loop, and in some cases with 3-4 Pis and batteries and related accessories, and have never had issues going through security because of my loop.)
+
+* **What do you do with the loop when you shower?**
+
Because the pumps aren't really waterproof, most of us choose to suspend and disconnect our pumps before we shower. You'll do the same thing even after you're looping. One trick, though, is to cancel any running temp basal rate and set a temp basal for 30 minutes with a rate of 0.0, and then suspend the pump. This will help OpenAPS accurately track your netIOB while you are off your pump. When you get out of the shower and are ready to reconnect your pump, do so. Make sure to unsuspend it. You can then either manually cancel the zero temp basal; or let OpenAPS read and decide what temp basal to issue next.
+
+* **What do you do when you exercise?**
+
This varies from person to person, and depends on the type and length of activity. Here's a few tidbits from [Dana](http://twitter.com/danamlewis) on how she does various activities. (Other loopers, PR into this page with your additional tips and how-to's.)
+ * **Hiking** - Definitely take the loop with! Think about setting a temporary target (you can enter it in Nightscout if you have connectivity) higher for the duration of the exercise. If you're offline, just change your targets in your pump. The loop will read the adjusted targets and begin looping toward that target. When you're done with the activity, change your targets back. In this scenario, I might change my loop target from 100 (normal day or nightime) to 130 or 140 as a target.
+ * **Swimming, Snorkeling, Scuba Diving, etc. (water sports)** - You can't loop while you're in the water, because the pump is not waterproof. (Unless you're sitting in a hot tub and have your pump safely above water, along with your CGM sensor being above water so it can transmit to the receiver, which is also not waterproof.) You can try having your sensor on your arm and keeping it above water so it can read every now and then if the receiver is in range. That being said, again, pump is NOT waterproof so you'll need to apply shower methodology (temp to zero, suspend, take pump off) to best track your netIOB. Some people observe having the CGM, once it gets back into range and reads data after the sensor has been submerged, read falsely high. It's not a big deal for the loop (because it's looking at trends, and doses using temp basals in a conservative way), but you'll likely want to fingerstick and/or wait a while before you'll be really happy with your CGM results again.
+
+ * **Running** - If it is a short run, (<30 minutes), I may not take the loop with me because any adjustments it would make are going to impact me after the run is done. For longer runs, I often now take my small, Edison based rig which can slip into the pocket of my hand-held running drink bottle that holds Gatorade. Before any length run, I try to make sure I don't have much positive netIOB on board (that's the biggest key to success). I also turn on activity mode (essentially a temp target of 120-140 or changing my pump targets to 120-140) an hour or so before a run and during the run; especially if I am carrying the loop during the run.
+
+ For any exercise or activity or time period, if you do not choose to take your loop (or if you forget it), the loop will pick up again once you get back into range and resume. (This is why it's important to temp then suspend so it can track the amount of insulin you haven't been getting.)
+
diff --git a/docs/docs/walkthrough/phase-6/advanced-features.md b/docs/docs/walkthrough/phase-6/advanced-features.md
index 8648b2d9e..24d44ae4a 100644
--- a/docs/docs/walkthrough/phase-6/advanced-features.md
+++ b/docs/docs/walkthrough/phase-6/advanced-features.md
@@ -11,3 +11,9 @@ Like all features and steps, you'll want to carefully enable, test, and observe
## Auto-sensitivity mode
Wouldn't it be great if the system knew when you were running sensitive or resistant? That's what we thought, so we created "auto-sensitivity mode". If you explicitly configure this additional feature, it will allow the system to analyze historical data on the go and make adjustments if it recognizes that you are reacting more sensitivite (or conversely, more resistant) to insulin than usual. It will then make micro adjustments to your basals.
+
+## Battery monitoring
+
+Because running OpenAPS requires frequent communication with your pump, your pump battery tends to drain more quickly than you'd experience when not looping. Some users have had good experiences with Energizer Ultimate Lithium AAA batteries (getting ~1.5weeks) rather than alkaline batteries (getting ~2-3 days). Regardless of whether you use alkaline or lithium, you may want to consider a Nightscout alarm to alert you to when the battery is running low. You can do this by setting (in your Nightscout config vars) `PUMP_WARN_BATT_V` to 1.39 (if using lithium batteries, as is most common) and adding `battery` to your `PUMP_FIELDS` so that voltage is displayed on your Nightscout site.
+
+The 1.39 voltage warning will give you many hours (reportedly ~8+) heads up that you will need to change your battery. If you don't change the battery in time and end up with a "low battery" warning on the pump, the pump will still function, but RF communications will be turned off and you will not be able to loop until you put a new battery in.
diff --git a/docs/reference/oref0/oref0-get-profile.md b/docs/reference/oref0/oref0-get-profile.md
index 8dd39a2eb..fa31ab976 100644
--- a/docs/reference/oref0/oref0-get-profile.md
+++ b/docs/reference/oref0/oref0-get-profile.md
@@ -1,4 +1,4 @@
# `oref0-get-profile`
## Help
-usage: [ 'node', '/usr/local/bin/oref0-get-profile' ] [] []
+usage: [ 'node', '/usr/local/bin/oref0-get-profile' ] [] []
diff --git a/docs/reference/oref0/oref0-mint-max-iob.md b/docs/reference/oref0/oref0-mint-max-iob.md
index 8a4cdf0d9..9d3cfe3ee 100644
--- a/docs/reference/oref0/oref0-mint-max-iob.md
+++ b/docs/reference/oref0/oref0-mint-max-iob.md
@@ -1,11 +1,11 @@
# `oref0-mint-max-iob`
## Help
- Usage:
-oref0-mint-max-iob max_iob [max_iob.json]
+Usage:
+oref0-mint-max-iob [preferences.json]
oref0-mint-max-iob help - this message
-Print a perfect max_iob.json.
+Print a perfect preferences.json.
Examples:
@@ -16,6 +16,3 @@ $ oref0-mint-max-iob 2
$ oref0-mint-max-iob 2 foo.json
max_iob 2 saved in foo.json
-bewest@hither:~/src/openaps/oref0$
-
-