-
Notifications
You must be signed in to change notification settings - Fork 307
FIX: Revise the reproducibility of CompCor masks #2130
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
FIX: Revise the reproducibility of CompCor masks #2130
Conversation
This comment has been minimized.
This comment has been minimized.
8609326 to
692b15a
Compare
692b15a to
37105c6
Compare
309640f to
39d0498
Compare
39d0498 to
e0baea4
Compare
6bc0480 to
748d17e
Compare
|
Ready for you @rciric |
|
The horrible contours on our subsampled dataset will be addressed by nipreps/niworkflows#544. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
👍 looks great
fmriprep/utils/confounds.py
Outdated
| sigma = 0.5 * (zooms / imgzooms) | ||
| if len(sigma) > 1: | ||
| sigma = tuple(sigma) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
is this check necessary? (will sigma ever be <= 1?)
|
Hello, Apologies for the late response. I agree that morphological erosion using an isotropic fixed-voxel-size structuring element would generalize poorly to new resolutions. Reading the Behzadi paper again, I’m somewhat confused about the decision to warp the data into functional space before applying the erosion, and the motivation for doing so is unclear to me. I completely agree with the decision to perform the erosion in anatomical space, and the adaptive kernel size is a great idea. Eroding using a low-threshold GM mask intuitively sounds like an improvement over conventional methods, but I do think that we should ideally examine the correlations of WM and CSF time series with whole-brain global signal when using the new approach to determine whether there are any major changes from the conventional approach. In particular, we would expect that limiting partial volume effects keeps the tissue-specific correlation with global signal reasonably low. This is of particular concern for those researchers who believe strongly that global mean signal should not be discarded (although I do not find myself in that camp). My recommendation for tCompCor is similar: the proposed approach sounds very reasonable, although I think it wouldn't hurt to more clearly characterise the noise ROI and the component time series correlations at some point. tCompCor appears to be used with somewhat less frequency than aCompCor. To verify, are the new aCompCor masks also going to define the regions over which the (global) mean WM and CSF signals are extracted? The literature is not entirely uniform in use of anatomical CompCor approaches. In addition to the original method from Behzadi and colleagues, a similar aCompCor methodology proposed by Muschelli and colleagues is used with considerable frequency. In particular, the aCompCor evaluated by our group (and by Parkes et al. to my knowledge) more closely matched the Muschelli approach. By computing separate decompositions for each compartment mask and for the union mask, fMRIPrep already supports the most significant difference between the approaches. It's not, however, immediately clear to me whether Muschelli and colleagues performed their mask erosions in native anatomical space or in a standard space, since the maps they used were provided by a combined segmentation-normalisation procedure from SPM:
As a potential aid for evaluating the masks we obtain, I'm including here for reference the subject-wise voxel inclusion frequency map for WM and CSF masks in the Muschelli study.
Also potentially of note, Power and colleagues performed their mask erosion using FreeSurfer. I'm not sure whether this erosion was performed in volumetric or surface space, and I only have a passing familiarity with FreeSurfer. In any case, there is substantial precedent for eroding masks in subject anatomical space. This approach was most similar to what our group did in the benchmarking paper, although we performed our erosions morphologically in volumetric space.
The description in a later work suggests to me that the erosion was likely also in volumetric space, although I again cannot say with confidence. This study also includes an example of eroded masks (corresponding to the deepest WM colour below, although I seem to recall that many of the example masks I saw from the WashU group were restricted to deeper voxels still.)
Overall, I agree with the premises of this revision, although I believe that further practical characterisation of fMRIPrep's CompCor procedure, potentially in the form of empirical correlations and voxel inclusion maps, would be welcome at some future stage. |
|
I don't think the FreeSurfer method described could apply to the surface, as there is no representation of white matter or CSF on the surface. It sounds like they used the aseg segmentation. |
|
Hi @rciric, happy to do this:
If I understand correctly and to say that this PR improves the situation, we should calculate the correlation of global signal with the time-series extracted from the different CompCor masks. Then, if these correlations are not significantly larger than those before this PR, we can say these new masks at least don't make worse masks for compcor. Is that right? |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This looks overall reasonable. I'm assuming the Gaussian blur parameters are just heuristic, but if there's a principled justification, we should probably list it explicitly in a comment.
| return out_file | ||
|
|
||
|
|
||
| def acompcor_masks(in_files, is_aseg=False, zooms=None): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I don't really like sticking major pieces of machinery into a utils module and having the interfaces be thin wrappers on a hidden function. I think at one point we were doing something like:
class AlgorithmInterface:
def _run_interface(self, runtime):
self._results = _algorithm(**self.inputs.get())
return runtime
def _algorithm(arg1, arg2):
...
return {"out1": ...}And the justification was that it would be easier to test a function than an interface. We're not testing this, and now it's being hidden away into an unassuming corner of the code.
I don't want to hold up the PR, but I would suggest we think through our organization and apply it consistently.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I don't want to hold up the PR, but I would suggest we think through our organization and apply it consistently.
Yup, this would be good. Let's make sure we find a time after the LTS. At first my only comment would be that sphinx does not take private functions by default (that's why this is public). That said, if when we meet we decide to have everything together, fine by me - this was kind of an arbitrary decision.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks for the review :)
| return out_file | ||
|
|
||
|
|
||
| def acompcor_masks(in_files, is_aseg=False, zooms=None): |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
I don't want to hold up the PR, but I would suggest we think through our organization and apply it consistently.
Yup, this would be good. Let's make sure we find a time after the LTS. At first my only comment would be that sphinx does not take private functions by default (that's why this is public). That said, if when we meet we decide to have everything together, fine by me - this was kind of an arbitrary decision.
|
The masks in reports look pretty bad. |
|
Pushed your previous commit to |
You mean for ds054 and ds210 (i.e., I have mostly tested with recon-all - that's why I haven't lent much attention to ds054 and ds210. That said, I'm surprised that the combined masks look so bad. To recapitulate: Before this PR we were probably representing the combined mask (as stated in the caption of the reportlet), and it never ever contained CSF. I think our CSF mask was so exclusive that very few voxels were in it (and hence, the empty take errors from numpy). - I say this just in case you think ds005's reports look awful too. Now, the masks with recon-all look pretty good. I've checked all of them after running ds114, and @Shotgunosine has also generated a good bunch (which looked good to me and @rciric will soon assess after his quals). Then, we have the |
|
You're right, I didn't look at ds005 because it takes so much longer to complete. Do the voxels near the edge of the brain mask occur in higher resolution images? Are they problematic? I assume they're classified as CSF, but I would worry you get GM signal bleed-over (possibly resulting in the high variance for tCompCor). |
Are you referring to pink (aCompCor) or blue (tCompCor)? I guess they are not pink because the new implementation is quite careful - if they are selected, the certainty of them being CSF is very high. Regarding tCompCor, this is expected (which does not mean good). I particularly asked @rciric about this, and he didn't seem too concerned. Please note that the original Behzadi paper DID NOT erode the brain mask (as we were doing), and also, their top percentile was calculated slice-wise (with veeery thick slices). My impression is that the original implementation of tCompCor wasn't great because of this. We may want to come back to the previous implementation, but I definitely see the new tCompCor masks picking up voxels in the brain stem and external parts of the commissures - which is a good thing, I believe. |
They show up in both. Fair enough. |
|
Okay, running ds054 and ds210 at their original resolution with |
This commit revises the implementation of CompCor masks in an attempt to make it closer to the original proposal and at the same time address some recurring problems of *fMRIPrep*'s tCompCor implementation. Finally, with the more careful resampling of prior knowledge from the anatomical scan, this refactor should also make the aCompCor components more run-to-run repeatable. aCompCor -------- The massaging of CompCor masks is now done in anatomical space where it is more precise, and a careful resampling to BOLD space follows. The implementation deviates from Behzadi et al. Their original implementation thresholded the CSF and the WM partial-volume masks at 0.99 (i.e., 99% of the voxel volume is filled with a particular tissue), and then binary eroded that 2 voxels: > Anatomical data were segmented into gray matter, white matter, > and CSF partial volume maps using the FAST algorithm available > in the FSL software package (Smith et al., 2004). Tissue partial > volume maps were linearly interpolated to the resolution of the > functional data series using AFNI (Cox, 1996). In order to form > white matter ROIs, the white matter partial volume maps were > thresholded at a partial volume fraction of 0.99 and then eroded by > two voxels in each direction to further minimize partial voluming > with gray matter. CSF voxels were determined by first thresholding > the CSF partial volume maps at 0.99 and then applying a threedimensional > nearest neighbor criteria to minimize multiple tissue > partial voluming. Since CSF regions are typically small compared > to white matter regions mask, erosion was not applied. This particular procedure is not generalizable to BOLD data with different voxel zooms as the mathematical morphology operations will be scaled by those. Also, from reading the excerpt above and the tCompCor description, I (@oesteban) believe that they always operated slice-wise given the large slice-thickness of their functional data. Instead, *fMRIPrep*'s implementation deviates from Behzadi's implementation on two aspects: * the masks are prepared in high-resolution, anatomical space and then projected into BOLD space; and, * instead of using binary erosion, a dilated GM map is generated -- thresholding the corresponding PV map at 0.05 (i.e., pixels containing at least 5% of GM tissue) and then subtracting that map from the CSF, WM and CSF+WM (combined) masks. This should be equivalent to eroding the masks, except that the erosion only happens at direct interfaces with GM. When the probseg maps provene from FreeSurfer's ``recon-all`` (i.e., they are discrete), binary maps are *transformed* into some sort of partial volume maps by means of a Gaussian smoothing filter with sigma adjusted by the size of the BOLD data. tCompCor -------- In the case of *tCompCor*, this commit removes the heavy erosion of the brain mask because 1) that wasn't part of the original proposal by Behzadi et al., and 2) the erosion was the potential source of errors from numpy complaining that it can't take from an empty axis of an array. > Based on these results, we chose a 2% threshold > (∼20–30 voxels per slice) as a reasonable empirical > threshold that effectively identified voxels with > the highest fractional variance of physiological noise. Although they do the calculation slice-wise, this commit rolls tCompCor back to calculate the 2% threshold on the whole brain mask. Resolves: nipreps#2129. References: nipreps#2052.
Co-authored-by: Chris Markiewicz <[email protected]>
309264b to
bafb714
Compare
Masks look okay (better with FS, but that's the drill). I'm building a new singularity image (which required cutting an RC of sMRIPrep) to test these masks including the fix from Chris to make the reportlet show the combined mask instead of the wm mask. As soon as I have both datasets run with the latest image I'll ask @rciric for his trained eye. Meanwhile, please have a look into #2245 - I think it would be worth pushing into 20.1.x to be able to do the correlation w/global-signal test. |
|
My plan is to quickly make a PR to add the CompCor time series to the bar
plot of correlations with the GS (and remove the components from there).
That if @rciric thinks it's a good idea.
Worst case scenario, the plots will become more readable.
…On Wed, Sep 2, 2020, 14:57 Chris Markiewicz ***@***.***> wrote:
@oesteban <https://github.com/oesteban> @rciric
<https://github.com/rciric> What's the status of this? I think we're very
close to a release, so let me know if I can help on anything.
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#2130 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AAESDRSHLSWESN4TUY4AXRDSDY6L7ANCNFSM4NGMUUGQ>
.
|
|
Is this PR ready to merge, then? |
|
Yes, I think that's safe
…On Wed, Sep 2, 2020, 17:06 Chris Markiewicz ***@***.***> wrote:
Is this PR ready to merge, then?
—
You are receiving this because you were mentioned.
Reply to this email directly, view it on GitHub
<#2130 (comment)>,
or unsubscribe
<https://github.com/notifications/unsubscribe-auth/AAESDRXJYUT22W32H2R5QNTSDZNOPANCNFSM4NGMUUGQ>
.
|
20.2.0 RC0 (September 04, 2020) With this third minor release series of 2020, the first *fMRIPrep LTS* (*long-term support*) is finally here! This release contains a number of bug-fixes and enhancements mostly related to easing the maintenance, anticipating patch-release breaking changes to ensure a longstanding LTS, and addressing some run-to-run repeatability problems of the CompCor implementation. Thank you for using *fMRIPrep*! If you encounter any issues with this release, please let us know by posting an issue on our GitHub page! A full list of changes can be found below. * FIX: Revise the reproducibility of CompCor masks (nipreps#2130) * FIX: Simplify transform aggregation in resampling, pass identity transforms for multi-echo cases (nipreps#2239) * FIX: Skip the T1w check if ``--anat-derivatives`` is provided. (nipreps#2201) * FIX: Storing ``--bids-filters`` within config file (nipreps#2177) * FIX: Revise multi-echo reference generation, permitting using SBRefs too (nipreps#1803) * FIX: FreeSurfer license manipulation & canary * ENH: Output CompCor masks if ``--debug compcor`` is passed (nipreps#2248) * ENH: Conform to BIDS Derivatives as of BIDS 1.4.0 (nipreps#2223) * ENH: Reuse config (nipreps#2240) * ENH: Save BOLD-anatomical transforms to derivatives folder (nipreps#2233) * ENH: Leverage BIDSLayout's ``database_path`` (nipreps#2203) * ENH: Add ``--no-tty`` option to ``fmriprep-docker.py`` (nipreps#2204) * ENH: Report number of echoes in BOLD summary. (nipreps#2184) * ENH: Ensure NiPype telemetry is just pinged once (nipreps#2168) * DOC: Update reference in "Refinement of Brain Mask" description (nipreps#2215) * DOC: List *TemplateFlow* templates that need to be prefetched (nipreps#2196) * DOC: Update references to https://github.com/nipreps (nipreps#2191) * DOC: Pin NiPype with new Sphinx extension syntax (nipreps#2092) * MAINT: Track nipreps#2252 from 20.1.x series (nipreps#2253) * MAINT: Silence PyBIDS warning by setting extension mode (nipreps#2250) * MAINT: Drop CircleCI docs build (nipreps#2247) * MAINT: Pin latest *NiPreps* (nipreps#2244) * MAINT: Update ``setup.cfg`` (flake8 and pytest) (nipreps#2183) * MAINT: Delete release-drafter (nipreps#2169) * MAINT: Track bug-fix release on the 20.1.x series (nipreps#2165) * MAINT: Remove auto-comment bot (nipreps#2166) * MAINT: Improve the questions on the bug-report template (nipreps#2158)
20.2.0rc1
With this third minor release series of 2020,
the first *fMRIPrep LTS* (*long-term support*) is finally here!
This release contains a number of bug-fixes and enhancements mostly
related to easing the maintenance, anticipating patch-release breaking
changes to ensure a longstanding LTS, and addressing some run-to-run
repeatability problems of the CompCor implementation.
.. admonition:: Long-Term Support (LTS)
*fMRIPrep* 20.2 LTS introduces the `long-term support program
<https://www.nipreps.org/devs/releases/#long-term-support-series>`__.
This LTS version will be kindly steered and maintained by
the group of Dr. Basile Pinsard and Prof. Pierre Bellec at
`CRIUGM <http://www.criugm.qc.ca/>`__, (Université de Montréal).
The LTS is planned for a window of 4 years of support (i.e., until
September 2024).
.. caution::
As with all minor version increments, working directories
from previous versions **should not be reused**.
Thank you for using *fMRIPrep*!
If you encounter any issues with this release, please let us know
by posting an issue on our GitHub page!
A full list of changes can be found below.
* FIX: Get missing ``probseg`` file from MNI152NLin2009cAsym (nipreps#2271)
* FIX: Restore ``--ignore t2w/flair`` options (nipreps#2260)
* FIX: Revise the reproducibility of *CompCor* masks (nipreps#2130)
* FIX: Simplify transform aggregation in resampling, pass identity transforms for multi-echo cases (nipreps#2239)
* FIX: Skip the T1w check if ``--anat-derivatives`` is provided. (nipreps#2201)
* FIX: Storing ``--bids-filters`` within config file (nipreps#2177)
* FIX: Revise multi-echo reference generation, permitting using SBRefs too (nipreps#1803)
* FIX: *FreeSurfer* license manipulation & canary
* ENH: Output CompCor masks if ``--debug compcor`` is passed (nipreps#2248)
* ENH: Conform to BIDS Derivatives as of BIDS 1.4.0 (nipreps#2223)
* ENH: Reuse config (nipreps#2240)
* ENH: Save BOLD-anatomical transforms to derivatives folder (nipreps#2233)
* ENH: Leverage BIDSLayout's ``database_path`` (nipreps#2203)
* ENH: Add ``--no-tty`` option to ``fmriprep-docker.py`` (nipreps#2204)
* ENH: Report number of echoes in BOLD summary. (nipreps#2184)
* ENH: Ensure *NiPype* telemetry is just pinged once (nipreps#2168)
* DOC: Add FAQ entry for using pre-indexed layouts (nipreps#2256)
* DOC: Update reference in "Refinement of Brain Mask" description (nipreps#2215)
* DOC: List *TemplateFlow* templates that need to be prefetched (nipreps#2196)
* DOC: Update references to https://github.com/nipreps (nipreps#2191)
* DOC: Pin *NiPype* with new Sphinx extension syntax (nipreps#2092)
* MAINT: Track nipreps#2269 and nipreps#2269, bug-fixes on the 20.1.x series
* MAINT: Remove derivatives from layout index ignores (nipreps#2258)
* MAINT: Track nipreps#2252 from 20.1.x series (nipreps#2253)
* MAINT: Silence *PyBIDS* warning by setting extension mode (nipreps#2250)
* MAINT: Drop CircleCI docs build (nipreps#2247)
* MAINT: Pin latest *NiPreps* (nipreps#2244)
* MAINT: Update ``setup.cfg`` (flake8 and pytest) (nipreps#2183)
* MAINT: Delete release-drafter (nipreps#2169)
* MAINT: Track bug-fix release on the 20.1.x series (nipreps#2165)
* MAINT: Remove auto-comment bot (nipreps#2166)
* MAINT: Improve the questions on the bug-report template (nipreps#2158)
.. admonition:: Author list for papers based on *fMRIPrep* 20.2 LTS series
As described in the `Contributor Guidelines
<https://www.nipreps.org/community/CONTRIBUTING/#recognizing-contributions>`__,
anyone listed as developer or contributor may write and submit manuscripts
about *fMRIPrep*.
To do so, please move the author(s) name(s) to the front of the following list:
Markiewicz, Christopher J. \ :sup:`1`\ ; Goncalves, Mathias \ :sup:`1`\ ; DuPre, Elizabeth \ :sup:`2`\ ; Kent, James D. \ :sup:`3`\ ; Salo, Taylor \ :sup:`4`\ ; Ciric, Rastko \ :sup:`1`\ ; Pinsard, Basile \ :sup:`5`\ ; Finc, Karolina \ :sup:`6`\ ; de la Vega, Alejandro \ :sup:`7`\ ; Feingold, Franklin \ :sup:`1`\ ; Tooley, Ursula A. \ :sup:`8`\ ; Benson, Noah C. \ :sup:`9`\ ; Urchs, Sebastian \ :sup:`2`\ ; Blair, Ross W. \ :sup:`1`\ ; Erramuzpe, Asier \ :sup:`10`\ ; Lurie, Daniel J. \ :sup:`11`\ ; Heinsfeld, Anibal S. \ :sup:`12`\ ; Jacoby, Nir \ :sup:`13`\ ; Jamison, Keith W. \ :sup:`14`\ ; Frederick, Blaise B. \ :sup:`15, 16`\ ; Valabregue, Romain \ :sup:`17`\ ; Sneve, Markus H. \ :sup:`18`\ ; Liem, Franz \ :sup:`19`\ ; Adebimpe, Azeez \ :sup:`20`\ ; Velasco, Pablo \ :sup:`21`\ ; Wexler, Joseph B. \ :sup:`1`\ ; Groen, Iris I. A. \ :sup:`22`\ ; Ma, Feilong \ :sup:`23`\ ; Amlien, Inge K. \ :sup:`18`\ ; Bellec, Pierre \ :sup:`5`\ ; Cieslak, Matthew \ :sup:`20`\ ; Devenyi, Grabriel A. \ :sup:`24`\ ; Ghosh, Satrajit S. \ :sup:`25, 26`\ ; Gomez, Daniel E. P. \ :sup:`27`\ ; Halchenko, Yaroslav O. \ :sup:`23`\ ; Isik, Ayse Ilkay \ :sup:`28`\ ; Moodie, Craig A. \ :sup:`1`\ ; Naveau, Mikaël \ :sup:`29`\ ; Rivera-Dompenciel, Adriana \ :sup:`3`\ ; Satterthwaite, Theodore D. \ :sup:`20`\ ; Sitek, Kevin R. \ :sup:`30`\ ; Stojić, Hrvoje \ :sup:`31`\ ; Thompson, William H. \ :sup:`1`\ ; Wright, Jessey \ :sup:`1`\ ; Ye, Zhifang \ :sup:`32`\ ; Gorgolewski, Krzysztof J. \ :sup:`1`\ ; Poldrack, Russell A. \ :sup:`1`\ ; Esteban, Oscar \ :sup:`33`\ .
Affiliations:
1. Department of Psychology, Stanford University
2. Montreal Neurological Institute, McGill University
3. Neuroscience Program, University of Iowa
4. Department of Psychology, Florida International University
5. SIMEXP Lab, CRIUGM, University of Montréal, Montréal, Canada
6. Centre for Modern Interdisciplinary Technologies, Nicolaus Copernicus University in Toruń
7. University of Texas at Austin
8. Department of Neuroscience, University of Pennsylvania, PA, USA
9. Department of Psychology, New York University
10. Computational Neuroimaging Lab, BioCruces Health Research Institute
11. Department of Psychology, University of California, Berkeley
12. Child Mind Institute
13. Department of Psychology, Columbia University
14. Department of Radiology, Weill Cornell Medicine
15. McLean Hospital Brain Imaging Center, MA, USA
16. Consolidated Department of Psychiatry, Harvard Medical School, MA, USA
17. CENIR, INSERM U1127, CNRS UMR 7225, UPMC Univ Paris 06 UMR S 1127, Institut du Cerveau et de la Moelle épinière, ICM, F-75013, Paris, France
18. Center for Lifespan Changes in Brain and Cognition, University of Oslo
19. URPP Dynamics of Healthy Aging, University of Zurich
20. Perelman School of Medicine, University of Pennsylvania, PA, USA
21. Center for Brain Imaging, New York University
22. Department of Psychology, New York University, NY, USA
23. Dartmouth College: Hanover, NH, United States
24. Department of Psychiatry, McGill University
25. McGovern Institute for Brain Research, MIT, MA, USA
26. Department of Otolaryngology, Harvard Medical School, MA, USA
27. Donders Institute for Brain, Cognition and Behaviour, Radboud University Nijmegen
28. Max Planck Institute for Empirical Aesthetics
29. Cyceron, UMS 3408 (CNRS - UCBN), France
30. Speech & Hearing Bioscience & Technology Program, Harvard University
31. Max Planck UCL Centre for Computational Psychiatry and Ageing Research, University College London
32. State Key Laboratory of Cognitive Neuroscience and Learning, Beijing Normal University
33. Department of Radiology, CHUV, Université de Lausanne
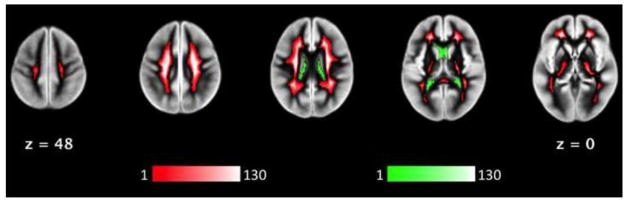
This PR revises the implementation of CompCor masks in an attempt to make it closer to the original proposal and at the same time address some recurring problems of fMRIPrep's tCompCor implementation.
Finally, with the more careful resampling of prior knowledge from the anatomical scan, this refactor should also make the aCompCor components more run-to-run repeatable.
aCompCor
The massaging of CompCor masks is now done in anatomical space where it is more precise, and a careful resampling to BOLD space follows.
The implementation deviates from Behzadi et al. Their original implementation thresholded the CSF and the WM partial-volume masks at 0.99 (i.e., 99% of the voxel volume is filled with a particular tissue), and then binary eroded that 2 voxels:
This particular procedure is not generalizable to BOLD data with different voxel zooms as the mathematical morphology operations will be scaled by those. Also, from reading the excerpt above and the tCompCor description, I (@oesteban)
believe that they always operated slice-wise given the large slice-thickness of their functional data.
Instead, fMRIPrep's implementation deviates from Behzadi's implementation on two aspects:
When the probseg maps provene from FreeSurfer's
recon-all(i.e., they are discrete), binary maps are transformed into some sort of partial volume maps by means of a Gaussian smoothing filter with sigma adjusted by the size of theBOLD data.
tCompCor
In the case of tCompCor, this commit removes the heavy erosion of the brain mask because 1) that wasn't part of the original proposal by Behzadi et al., and 2) the erosion was the potential source of errors from numpy complaining that it can't take from an empty axis of an array.
Although they do the calculation slice-wise, this commit rolls tCompCor back to calculate the 2% threshold on the whole-brain mask.
I would appreciate a deep review from @rciric & @jdkent to ensure my interpretation of Behzadi's paper is correct. I'm also tagging @effigies for a more top-level check of the changes. Further feedback from others is of course very much appreciated.
Resolves: #2129.
References: #2052.