-
-
Notifications
You must be signed in to change notification settings - Fork 81
Closed
Description
Creating here separate issue to keep track of stuff in addition to #394,
I think it's useful to know which reactions form subnetworks in the reaction network based on linkage class.
"""
Given a `ReactionSystem` , returns a Vector of Vector of Reactions that form
subnetworks based on linkage class
"""
function subnetworks( rs::ReactionSystem;sparse::Bool=false,
rmap::Vector{Vector{Pair{Int64, Int64}}} = collect(values(reactioncomplexmap(rs))),
r::Vector{Reaction} = reactions(rs),
ig::SimpleDiGraph{Int64} = incidencematgraph(reactioncomplexes(rs;sparse=sparse)[2]),
lc::Vector{Vector{Int64}} = linkageclasses(ig) )
subrs = Vector{Vector{Reaction}}()
for i in 1:length(lc)
rxind = unique(vcat([map(first, rmap[lc[i]][j]) for j in 1:length(lc[i])]...))
push!(subrs , r[rxind])
end
subrs
end
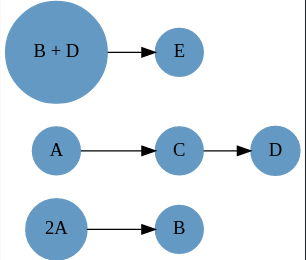
rn = @reaction_network begin
k₁, 2A --> B
k₂, A --> C
k₃, C --> D
k₄, B + D --> E
end k₁ k₂ k₃ k₄
julia> subnetworks(rn)
3-element Vector{Vector{Reaction}}:
[Reaction{Any, Int64}: k₁, 2A --> B]
[Reaction{Any, Int64}: k₂, A --> C, Reaction{Any, Int64}: k₃, C --> D]
[Reaction{Any, Int64}: k₄, B + D --> E]Implying there are three subnetworks that do not share reaction-complexes.. This is also evident from
complexgraph(rn)Metadata
Metadata
Assignees
Labels
No labels